Comparing predicted UTRs
Philipp Ross
2018-09-25
Last updated: 2018-11-18
workflowr checks: (Click a bullet for more information)-
✖ R Markdown file: uncommitted changes
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can runwflow_publishto commit the R Markdown file and build the HTML. -
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(12345)The command
set.seed(12345)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: dd9d56a
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/.DS_Store Ignored: analysis/.httr-oauth Ignored: analysis/figure/ Ignored: code/.DS_Store Ignored: code/differential_expression/ Ignored: code/differential_phase/ Ignored: data/ Ignored: docs/.DS_Store Ignored: docs/figure/.DS_Store Ignored: docs/figure/neighboring_genes.Rmd/.DS_Store Ignored: output/compare/ Ignored: output/ctss_clustering/ Ignored: output/differential_detection/ Ignored: output/differential_expression/ Ignored: output/differential_phase/ Ignored: output/extensive_transcription/ Ignored: output/final_utrs/ Ignored: output/gcbias/ Ignored: output/homopolymer_analysis/ Ignored: output/neighboring_genes/ Ignored: output/promoter_architecture/ Ignored: output/tfbs_analysis/ Ignored: output/transcript_abundance/ Untracked files: Untracked: _workflowr.yml Untracked: docs/figure/tfbs_analysis.Rmd/ Untracked: figures/ Unstaged changes: Modified: README.md Modified: analysis/_site.yml Modified: analysis/about.Rmd Modified: analysis/analyze_neighboring_genes.Rmd Modified: analysis/array_correlations.Rmd Modified: analysis/calculate_transcript_abundance.Rmd Deleted: analysis/chunks.R Modified: analysis/comparing_utrs.Rmd Modified: analysis/ctss_clustering.Rmd Modified: analysis/dynamic_tss.Rmd Modified: analysis/extensive_transcription.Rmd Modified: analysis/final_utrs.Rmd Modified: analysis/gcbias.Rmd Modified: analysis/index.Rmd Modified: analysis/license.Rmd Modified: analysis/process_neighboring_genes.Rmd Modified: analysis/promoter_architecture.Rmd Modified: analysis/strain_differential_detection.Rmd Modified: analysis/strain_differential_expression.Rmd Modified: analysis/strain_differential_phase.Rmd Modified: analysis/tfbs_analysis.Rmd Modified: code/differential_detection/detect_transcripts.R Deleted: docs/Rplots.pdf
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 3e6a944 | Philipp Ross | 2018-09-25 | updated utr comparison |
| html | 3e6a944 | Philipp Ross | 2018-09-25 | updated utr comparison |
| html | c5562e7 | Philipp Ross | 2018-09-25 | updated utr comparison |
| Rmd | fa4fca8 | Philipp Ross | 2018-09-25 | added homopolymer analysis |
| Rmd | 1e6d9bb | Philipp Ross | 2018-09-24 | comparing UTRs |
| html | 1e6d9bb | Philipp Ross | 2018-09-24 | comparing UTRs |
Comparing different UTR and TSS predictions
Since we were able to predict both UTRs and TSSs using our data, we wanted to know how our predictions compared to previously published predictions. Here, we compare the UTRs predicted in Caro et al. and Adjalley et al..
What we see is that although there are some large deviations, for the majority of 5UTR and TSS predictions, the results are not very different with a mean hovering around zero base pairs of difference between the start positions of our predicted 5’ UTRs and TSS and those that were previously published.
First, let’s import our 5UTR data:
Comparing to Caro et al.
# Comparing our UTR estimates to the Derisi predictions
derisi1 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410291_UTRs_1.bed"))
derisi2 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410292_UTRs_2.bed"))
derisi3 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410293_UTRs_3.bed"))
derisi4 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410294_UTRs_4.bed"))
derisi5 <- tibble::as_tibble(rtracklayer::import.bed("../data/compare_utrs/GSM1410295_UTRs_5.bed"))
fix_derisi_utrs <- function(set) {
set$name <- stringi::stri_replace_last(set$name,replacement=" ",regex="_")
set <- set %>% tidyr::separate(name,into = c("gene_id","type"), sep =" ")
set$gene_id <- toupper(set$gene_id)
out <- dplyr::inner_join(set, aliases, by=c("gene_id"="alias_symbol"))
return(out)
}
derisi1 <- fix_derisi_utrs(derisi1)
derisi2 <- fix_derisi_utrs(derisi2)
derisi3 <- fix_derisi_utrs(derisi3)
derisi4 <- fix_derisi_utrs(derisi4)
derisi5 <- fix_derisi_utrs(derisi5)Timepoint 1
tmp1 <- dplyr::select(derisi1, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
g <- ggplot(compare_derisi,aes(width.x,width.y)) +
geom_point() +
geom_abline(color="red") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_scatter_1.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_scatter_1.svg",plot=g)
print(g)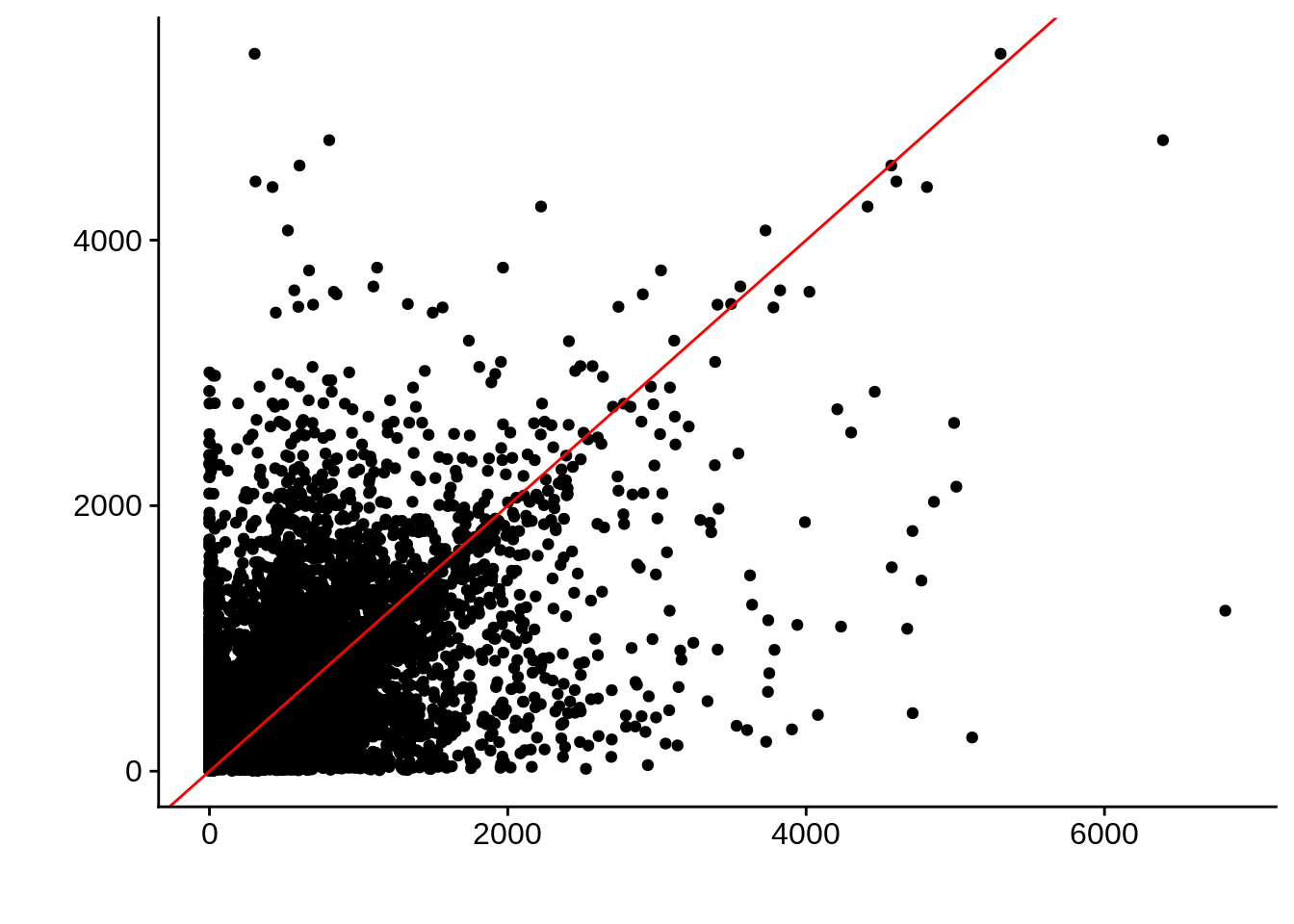
Expand here to see past versions of unnamed-chunk-4-1.png:
| Version | Author | Date |
|---|---|---|
| 3e6a944 | Philipp Ross | 2018-09-25 |
| 1e6d9bb | Philipp Ross | 2018-09-24 |
g <- ggplot(compare_derisi,aes(width.x-width.y)) +
geom_histogram(color="grey70") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_histogram_1.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_histogram_1.svg",plot=g)
print(g)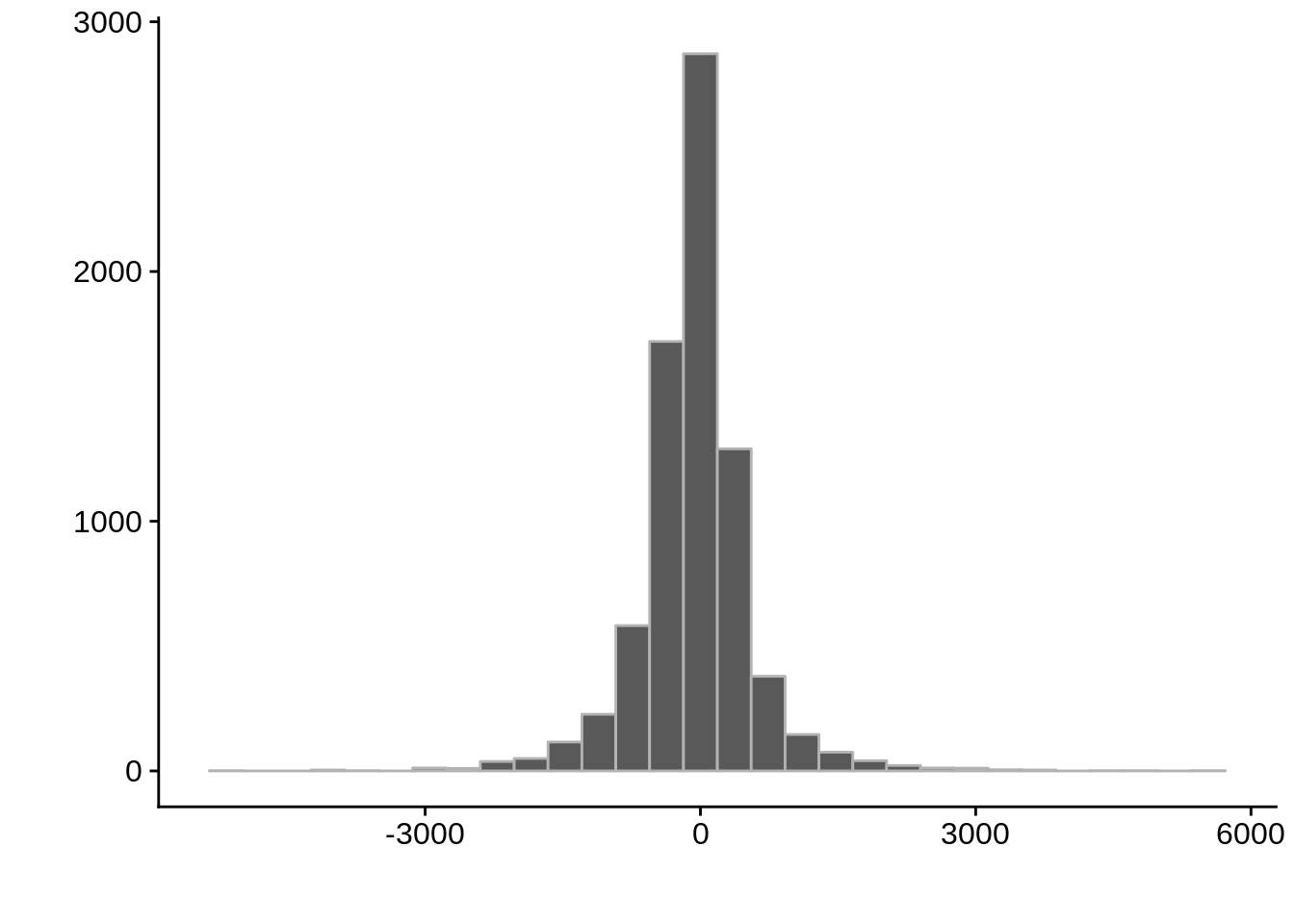
Expand here to see past versions of unnamed-chunk-4-2.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
Pearson's product-moment correlation
data: compare_derisi$width.x and compare_derisi$width.y
t = 44.624, df = 7619, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.4372115 0.4728149
sample estimates:
cor
0.4551951 gene_id.y width.x width.y diff
Length:7621 Min. : 0.0 Min. : 2 Min. :-5101.00
Class :character 1st Qu.: 221.0 1st Qu.: 281 1st Qu.: -328.00
Mode :character Median : 498.0 Median : 513 Median : -37.00
Mean : 610.5 Mean : 674 Mean : -63.45
3rd Qu.: 810.0 3rd Qu.: 895 3rd Qu.: 196.00
Max. :6810.0 Max. :5404 Max. : 5601.00 Timepoint 2
tmp1 <- dplyr::select(derisi2, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
g <- ggplot(compare_derisi,aes(width.x,width.y)) +
geom_point() +
geom_abline(color="red") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_scatter_2.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_scatter_2.svg",plot=g)
print(g)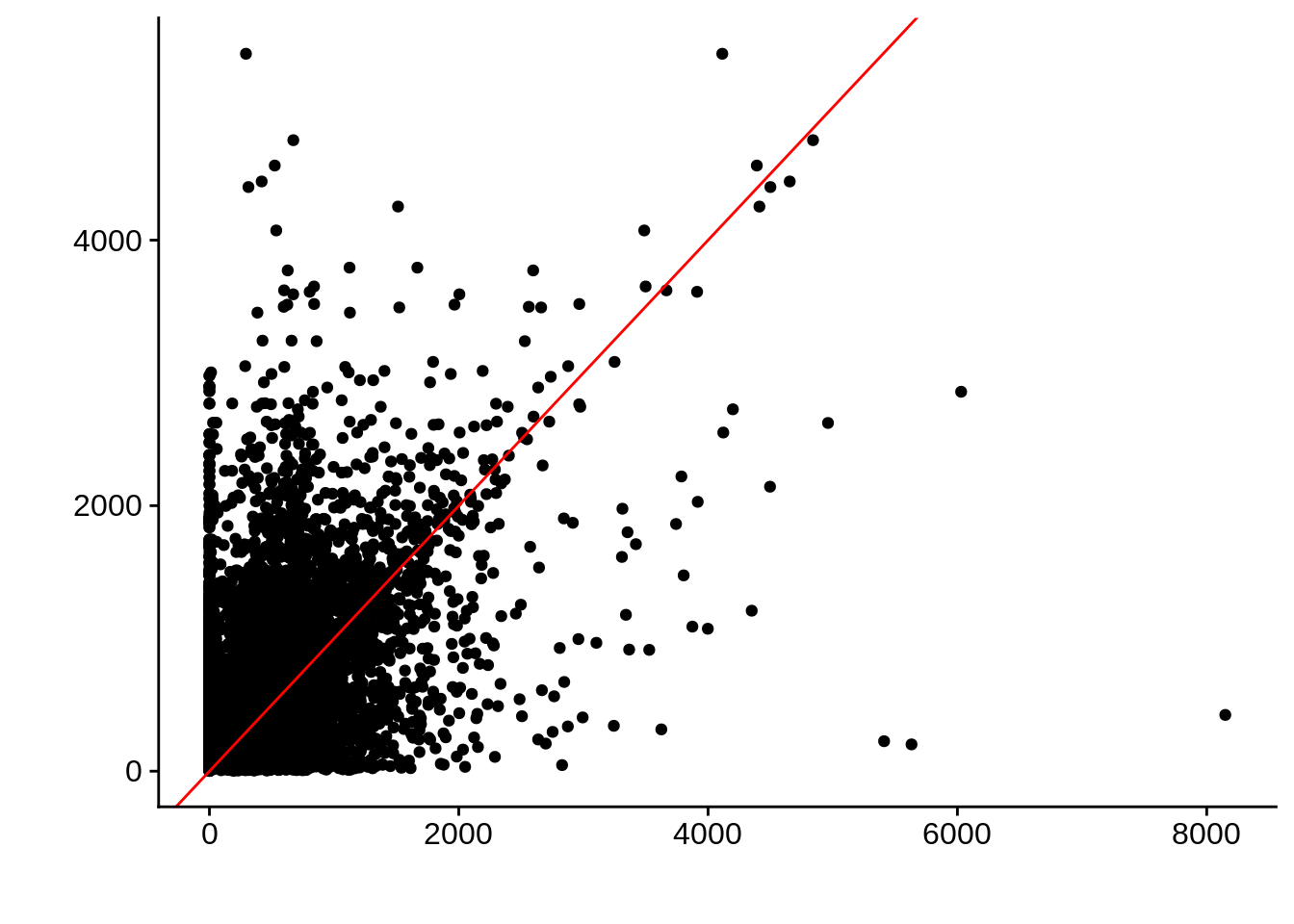
Expand here to see past versions of unnamed-chunk-5-1.png:
| Version | Author | Date |
|---|---|---|
| 3e6a944 | Philipp Ross | 2018-09-25 |
| 1e6d9bb | Philipp Ross | 2018-09-24 |
g <- ggplot(compare_derisi,aes(width.x-width.y)) +
geom_histogram(color="grey70") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_histogram_2.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_histogram_2.svg",plot=g)
print(g)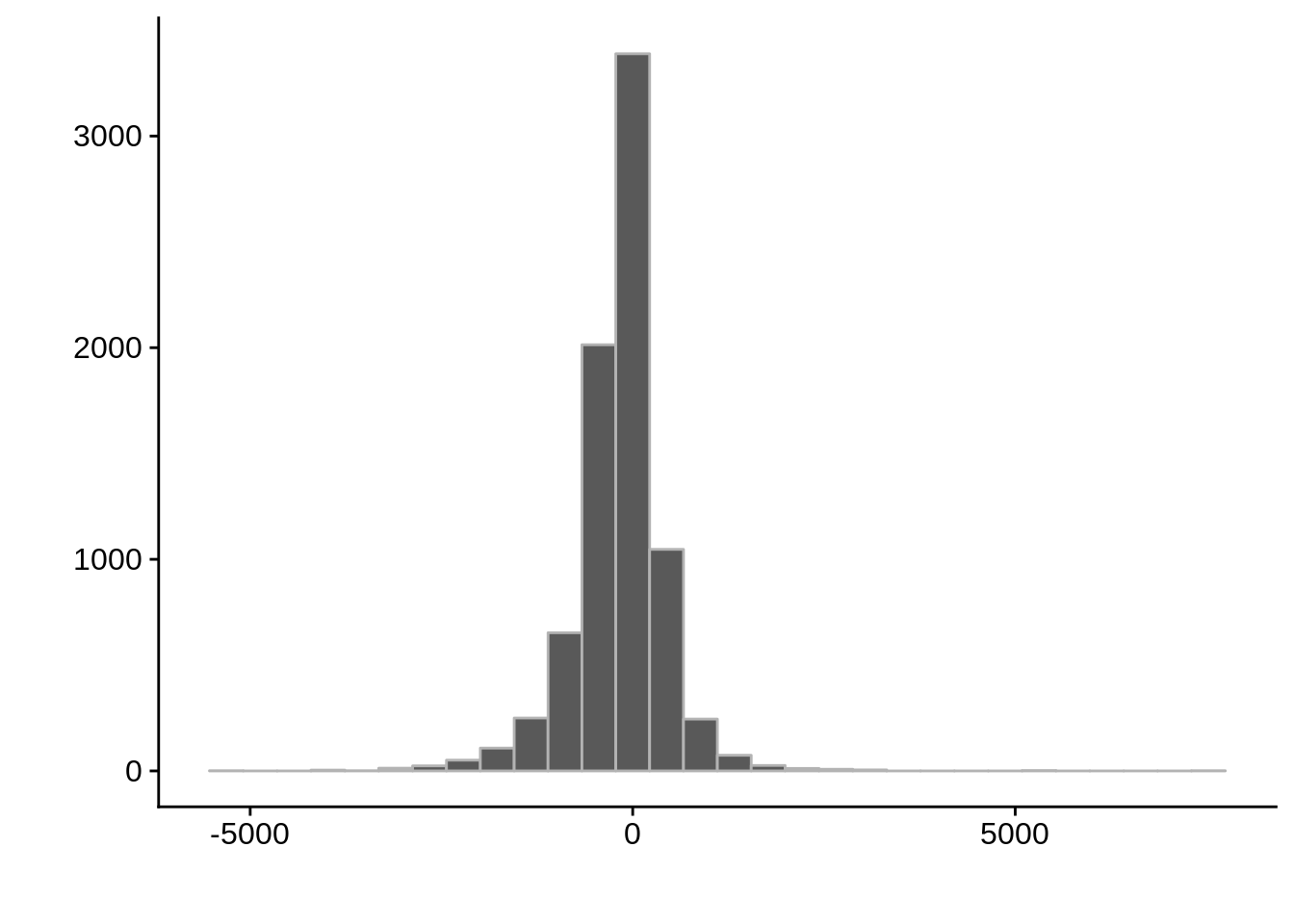
Expand here to see past versions of unnamed-chunk-5-2.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
Pearson's product-moment correlation
data: compare_derisi$width.x and compare_derisi$width.y
t = 39.839, df = 7926, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.3899543 0.4266379
sample estimates:
cor
0.408461 gene_id.y width.x width.y diff
Length:7928 Min. : 0.0 Min. : 2.0 Min. :-5111.0
Class :character 1st Qu.: 146.0 1st Qu.: 283.0 1st Qu.: -418.0
Mode :character Median : 428.0 Median : 518.0 Median : -93.0
Mean : 508.4 Mean : 675.7 Mean : -167.4
3rd Qu.: 690.2 3rd Qu.: 897.0 3rd Qu.: 120.0
Max. :8149.0 Max. :5404.0 Max. : 7725.0 Timepoint 3
tmp1 <- dplyr::select(derisi3, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
g <- ggplot(compare_derisi,aes(width.x,width.y)) +
geom_point() +
geom_abline(color="red") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_scatter_3.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_scatter_3.svg",plot=g)
print(g)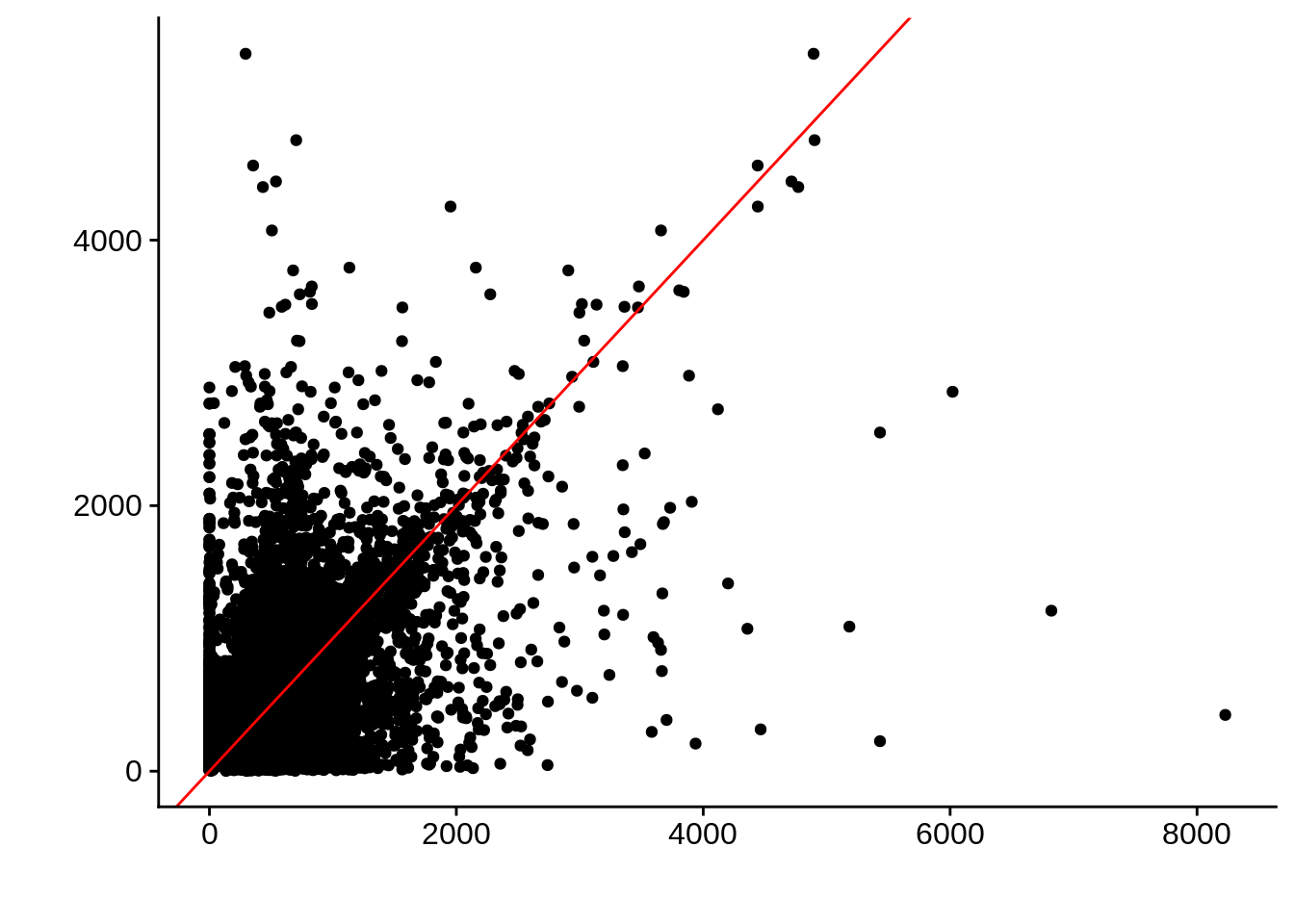
Expand here to see past versions of unnamed-chunk-6-1.png:
| Version | Author | Date |
|---|---|---|
| 3e6a944 | Philipp Ross | 2018-09-25 |
| 1e6d9bb | Philipp Ross | 2018-09-24 |
g <- ggplot(compare_derisi,aes(width.x-width.y)) +
geom_histogram(color="grey70") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_histogram_3.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_histogram_3.svg",plot=g)
print(g)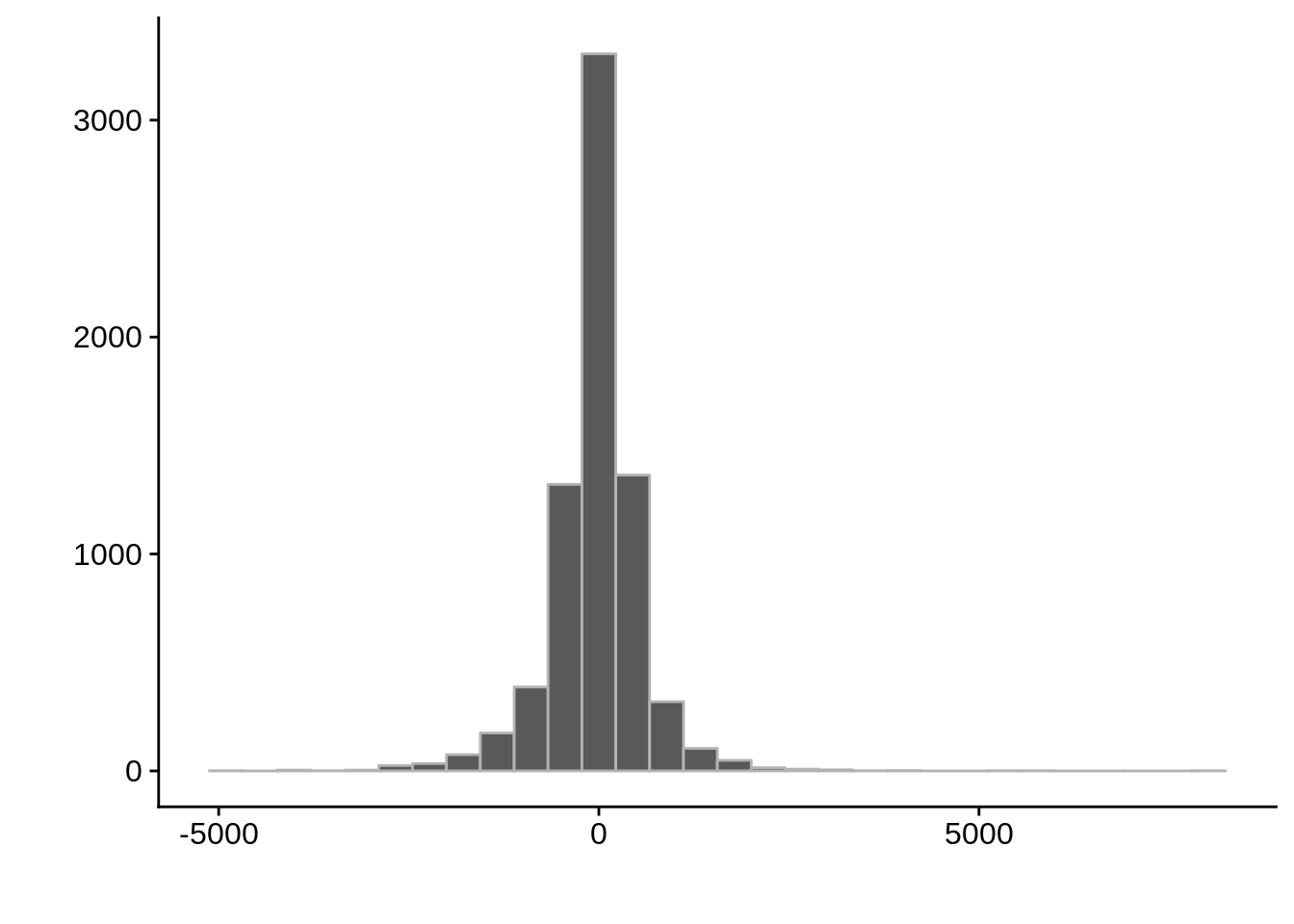
Expand here to see past versions of unnamed-chunk-6-2.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
Pearson's product-moment correlation
data: compare_derisi$width.x and compare_derisi$width.y
t = 40.612, df = 7201, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.4127160 0.4503005
sample estimates:
cor
0.4316956 gene_id.y width.x width.y diff
Length:7203 Min. : 0.0 Min. : 2 Min. :-5111.0
Class :character 1st Qu.: 346.5 1st Qu.: 289 1st Qu.: -268.0
Mode :character Median : 541.0 Median : 528 Median : 6.0
Mean : 655.6 Mean : 689 Mean : -33.4
3rd Qu.: 816.5 3rd Qu.: 912 3rd Qu.: 233.5
Max. :8229.0 Max. :5404 Max. : 7805.0 Timepoint 4
tmp1 <- dplyr::select(derisi4, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
g <- ggplot(compare_derisi,aes(width.x,width.y)) +
geom_point() +
geom_abline(color="red") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_scatter_4.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_scatter_4.svg",plot=g)
print(g)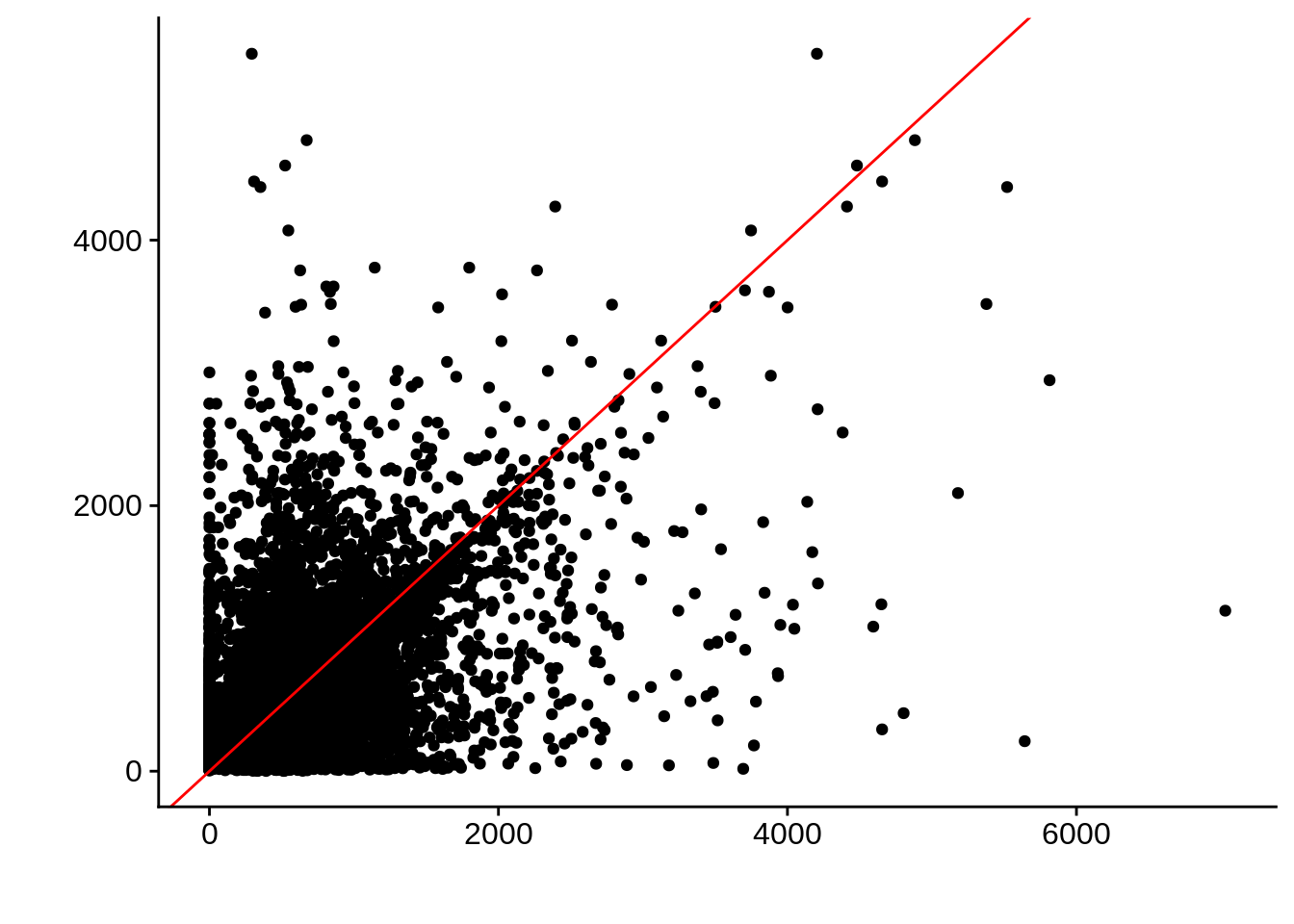
Expand here to see past versions of unnamed-chunk-7-1.png:
| Version | Author | Date |
|---|---|---|
| 3e6a944 | Philipp Ross | 2018-09-25 |
| 1e6d9bb | Philipp Ross | 2018-09-24 |
g <- ggplot(compare_derisi,aes(width.x-width.y)) +
geom_histogram(color="grey70") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_histogram_4.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_histogram_4.svg",plot=g)
print(g)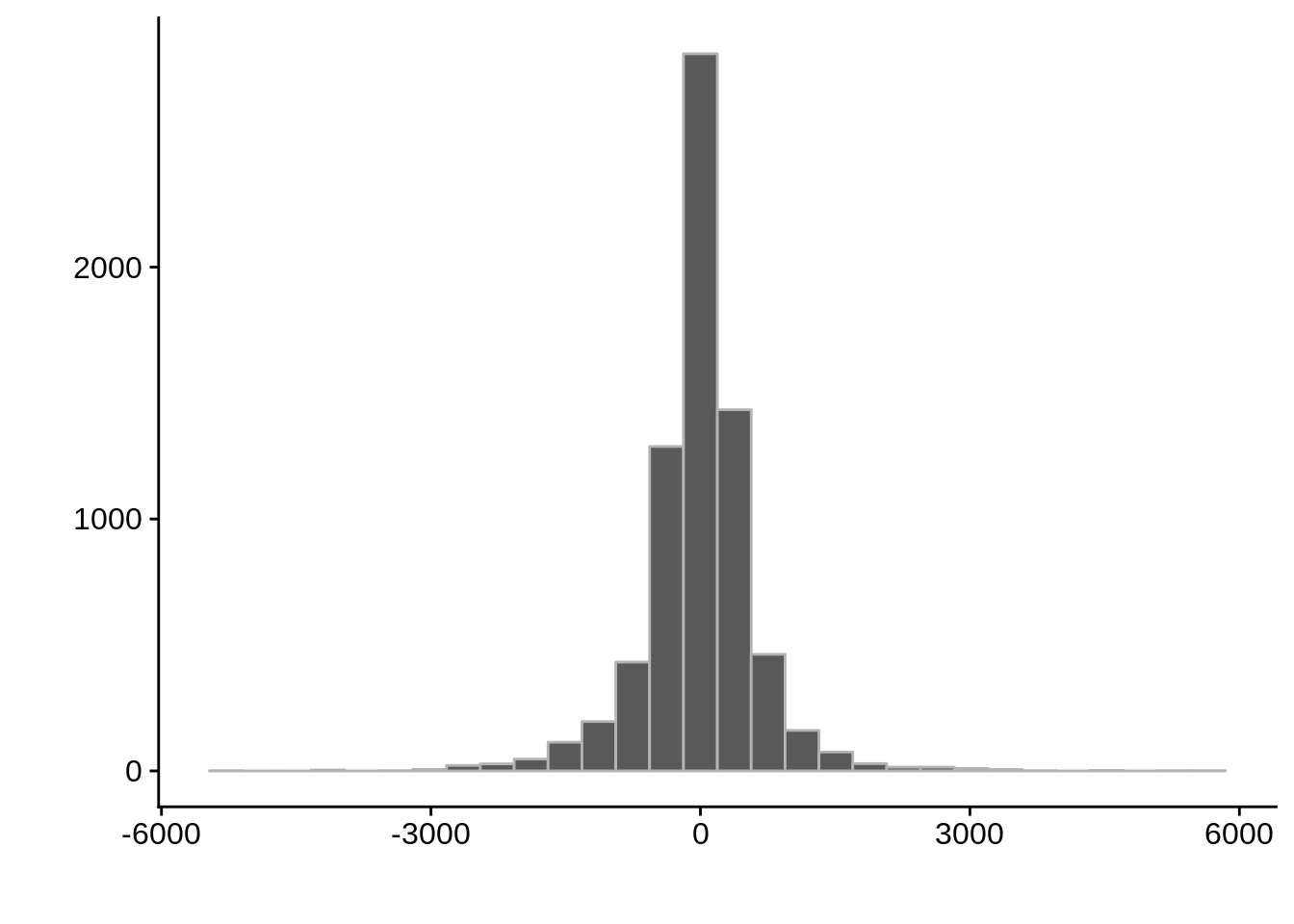
Expand here to see past versions of unnamed-chunk-7-2.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
Pearson's product-moment correlation
data: compare_derisi$width.x and compare_derisi$width.y
t = 39.132, df = 7197, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.3996269 0.4377261
sample estimates:
cor
0.4188608 gene_id.y width.x width.y diff
Length:7199 Min. : 0.0 Min. : 2.0 Min. :-5111.000
Class :character 1st Qu.: 340.0 1st Qu.: 287.0 1st Qu.: -261.500
Mode :character Median : 549.0 Median : 523.0 Median : 15.000
Mean : 674.9 Mean : 682.7 Mean : -7.868
3rd Qu.: 855.5 3rd Qu.: 902.0 3rd Qu.: 262.500
Max. :7030.0 Max. :5404.0 Max. : 5821.000 Timepoint 5
tmp1 <- dplyr::select(derisi5, gene_id.y, width)
tmp2 <- dplyr::select(utrs, Parent, width)
compare_derisi <- dplyr::inner_join(tmp1,tmp2,by=c("gene_id.y"="Parent"))
rm(tmp1,tmp2)
g <- ggplot(compare_derisi,aes(width.x,width.y)) +
geom_point() +
geom_abline(color="red") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_scatter_5.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_scatter_5.svg",plot=g)
print(g)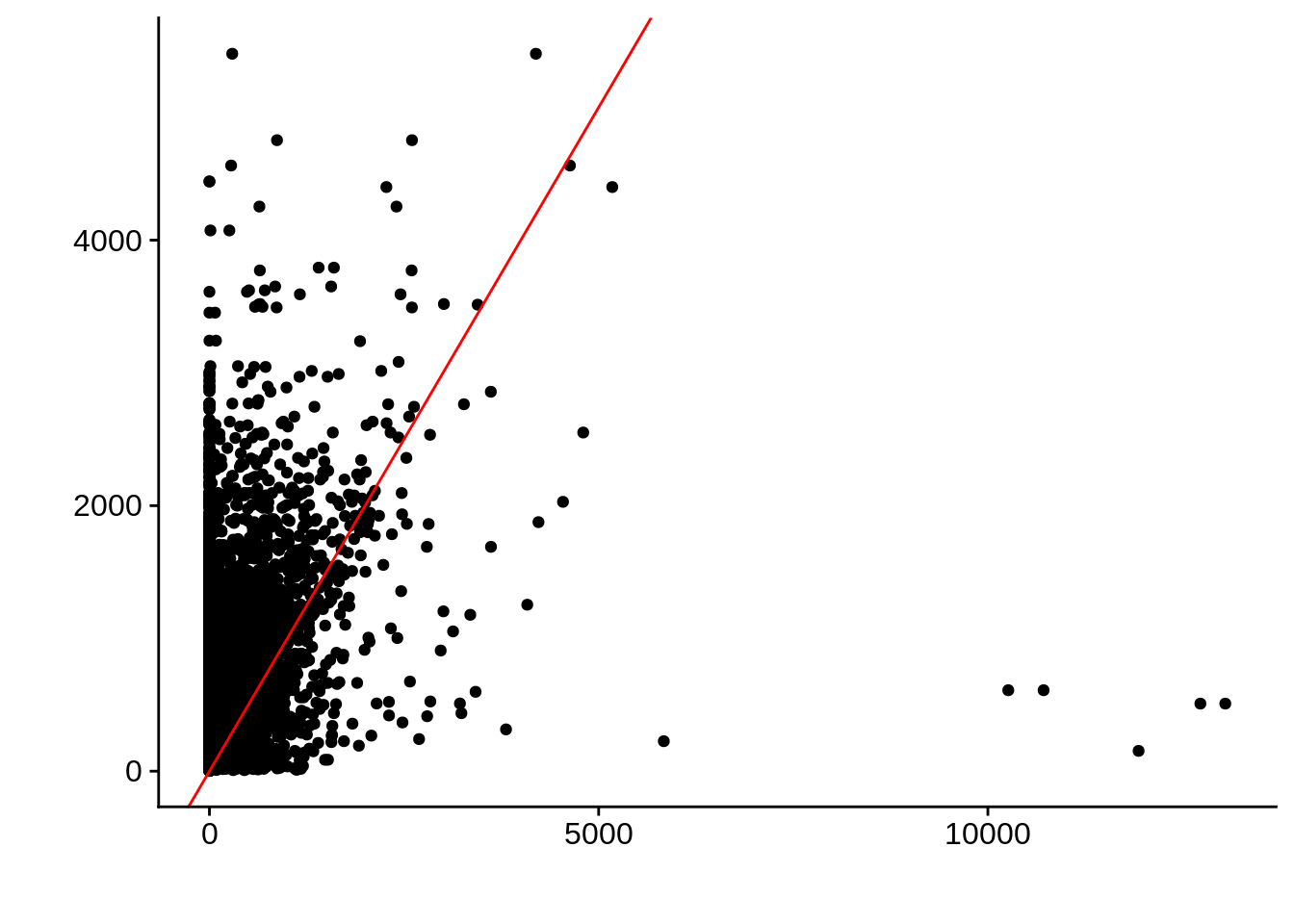
Expand here to see past versions of unnamed-chunk-8-1.png:
| Version | Author | Date |
|---|---|---|
| 3e6a944 | Philipp Ross | 2018-09-25 |
| 1e6d9bb | Philipp Ross | 2018-09-24 |
g <- ggplot(compare_derisi,aes(width.x-width.y)) +
geom_histogram(color="grey70") +
xlab("") +
ylab("")
cowplot::save_plot(filename="../output/compare/derisi_histogram_5.png",plot=g)
cowplot::save_plot(filename="../output/compare/derisi_histogram_5.svg",plot=g)
print(g)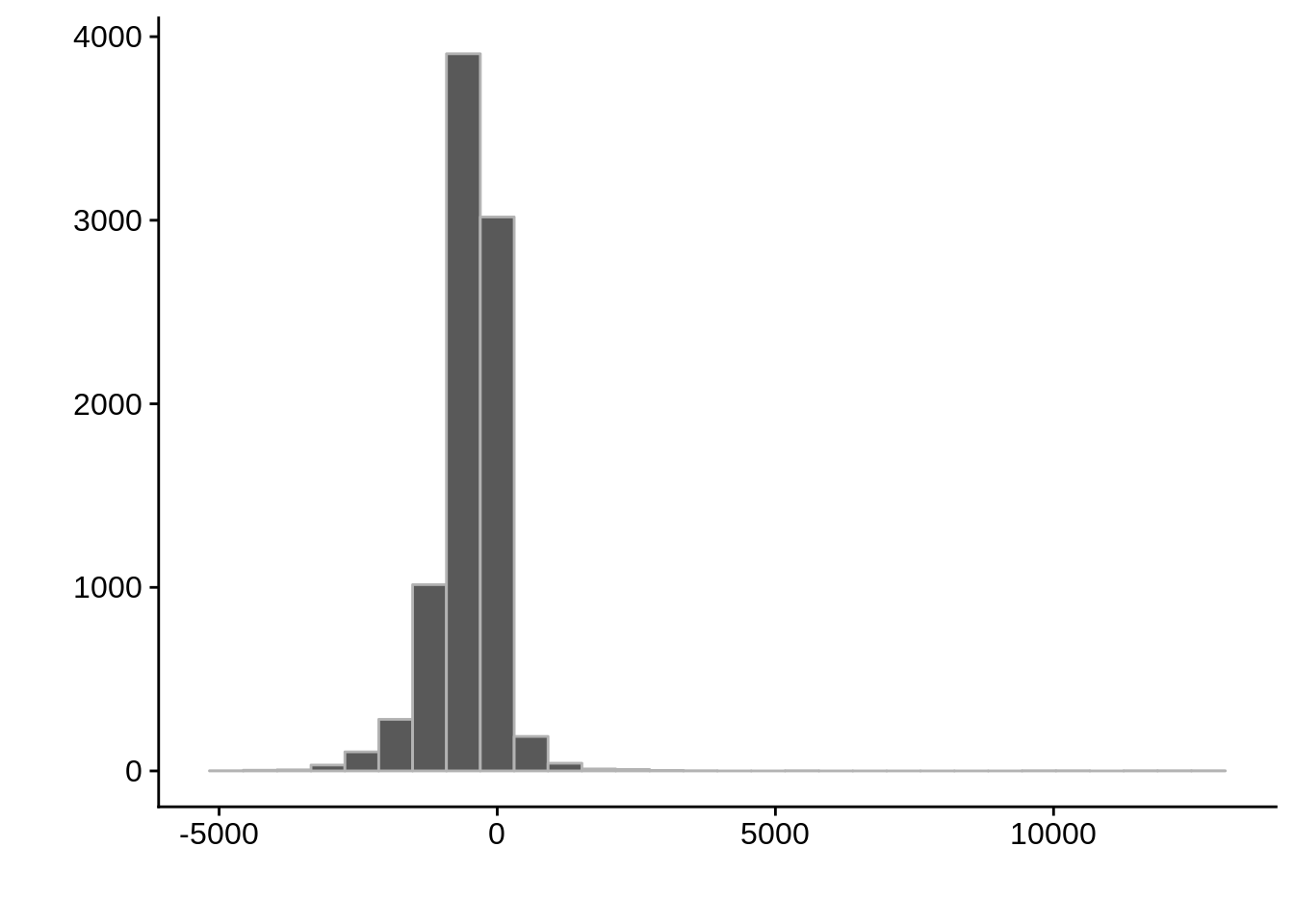
Expand here to see past versions of unnamed-chunk-8-2.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
Pearson's product-moment correlation
data: compare_derisi$width.x and compare_derisi$width.y
t = 29.152, df = 8625, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.2801573 0.3185779
sample estimates:
cor
0.299489 gene_id.y width.x width.y diff
Length:8627 Min. : 0.0 Min. : 2.0 Min. :-5111.0
Class :character 1st Qu.: 0.0 1st Qu.: 277.0 1st Qu.: -718.0
Mode :character Median : 0.0 Median : 508.0 Median : -415.0
Mean : 164.4 Mean : 660.9 Mean : -496.4
3rd Qu.: 63.0 3rd Qu.: 877.0 3rd Qu.: -189.0
Max. :13050.0 Max. :5404.0 Max. :12541.0 Comparing to Adjalley et al.
# Comparing our UTR estimates to the Adjalley predictions
adjalley <- tibble::as_tibble(rtracklayer::import.gff("../data/compare_utrs/sorted_Adjalley_Chabbert_TSSs.gff"))
for (i in 1:8) {
adjalley_filtered <- adjalley %>%
dplyr::mutate(position=(start+end)/2) %>%
dplyr::filter(FilterSize>i)
tmp1 <- dplyr::select(adjalley_filtered, AssignedFeat, position)
tmp2 <- dplyr::select(utrs, Parent, start)
compare_adjalley <- dplyr::inner_join(tmp1,tmp2,by=c("AssignedFeat"="Parent"))
compare_adjalley %>% dplyr::mutate(diff=position-start) %>% summary() %>% print()
g <- ggplot(compare_adjalley,aes(position-start)) +
geom_histogram(color="grey70") +
xlab("") +
ylab("")
cowplot::save_plot(filename=paste0("../output/compare/adjalley_histogram_",i,".png"),plot=g)
cowplot::save_plot(filename=paste0("../output/compare/adjalley_histogram_",i,".svg"),plot=g)
print(g)
} AssignedFeat position start diff
Length:36663 Min. : 40331 Min. : 43094 Min. :-4992.0
Class :character 1st Qu.: 428840 1st Qu.: 427513 1st Qu.: -222.0
Mode :character Median : 836668 Median : 836849 Median : 292.0
Mean : 963933 Mean : 963522 Mean : 410.9
3rd Qu.:1305788 3rd Qu.:1305295 3rd Qu.: 1044.0
Max. :3253500 Max. :3253655 Max. :12155.0 AssignedFeat position start diff
Length:29055 Min. : 41434 Min. : 43094 Min. :-4967.0
Class :character 1st Qu.: 421766 1st Qu.: 421138 1st Qu.: -262.0
Mode :character Median : 824344 Median : 823912 Median : 328.5
Mean : 947811 Mean : 947361 Mean : 449.5
3rd Qu.:1282446 3rd Qu.:1282646 3rd Qu.: 1169.5
Max. :3236321 Max. :3231222 Max. :12155.0 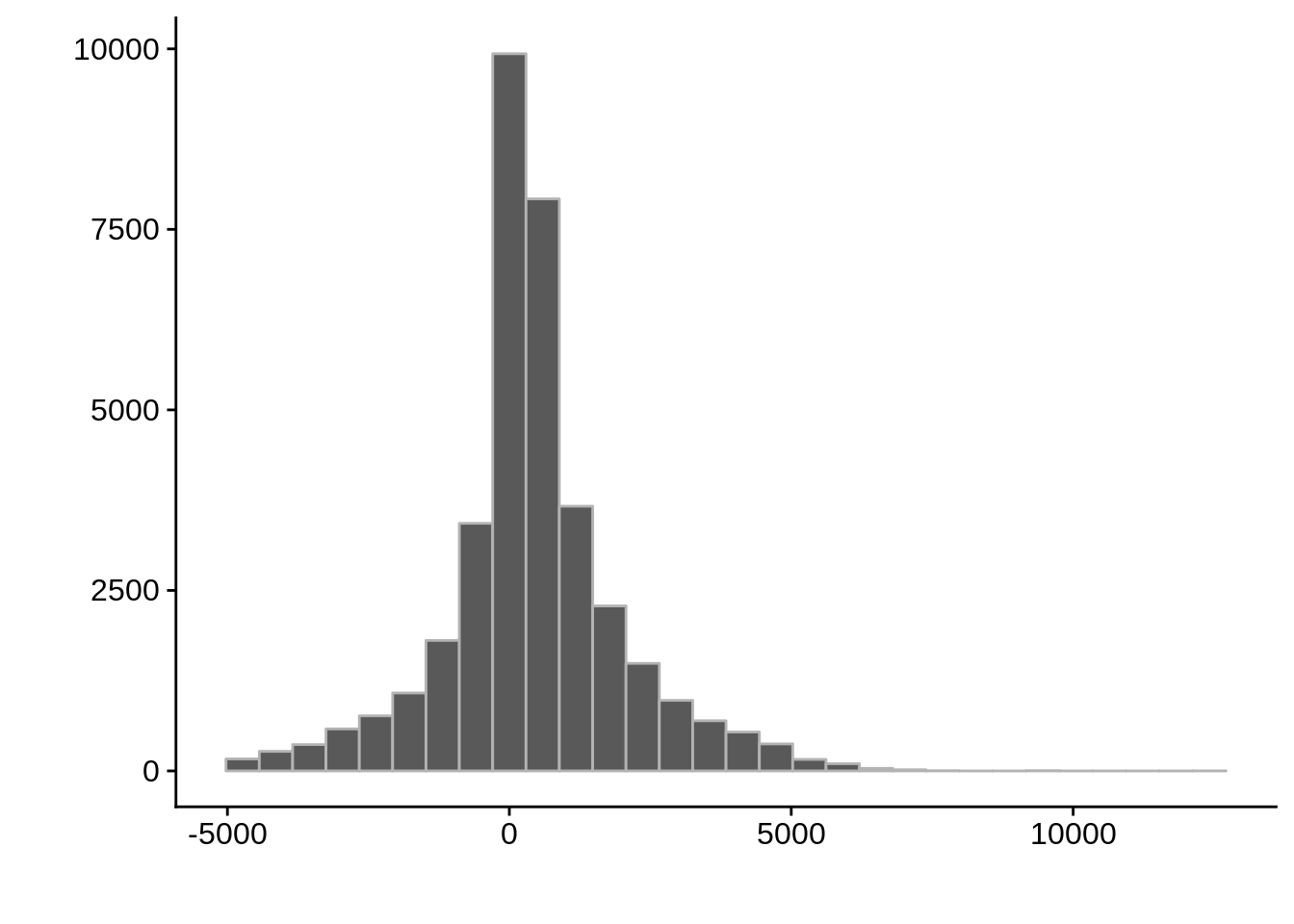
Expand here to see past versions of unnamed-chunk-9-1.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
AssignedFeat position start diff
Length:29055 Min. : 41434 Min. : 43094 Min. :-4967.0
Class :character 1st Qu.: 421766 1st Qu.: 421138 1st Qu.: -262.0
Mode :character Median : 824344 Median : 823912 Median : 328.5
Mean : 947811 Mean : 947361 Mean : 449.5
3rd Qu.:1282446 3rd Qu.:1282646 3rd Qu.: 1169.5
Max. :3236321 Max. :3231222 Max. :12155.0 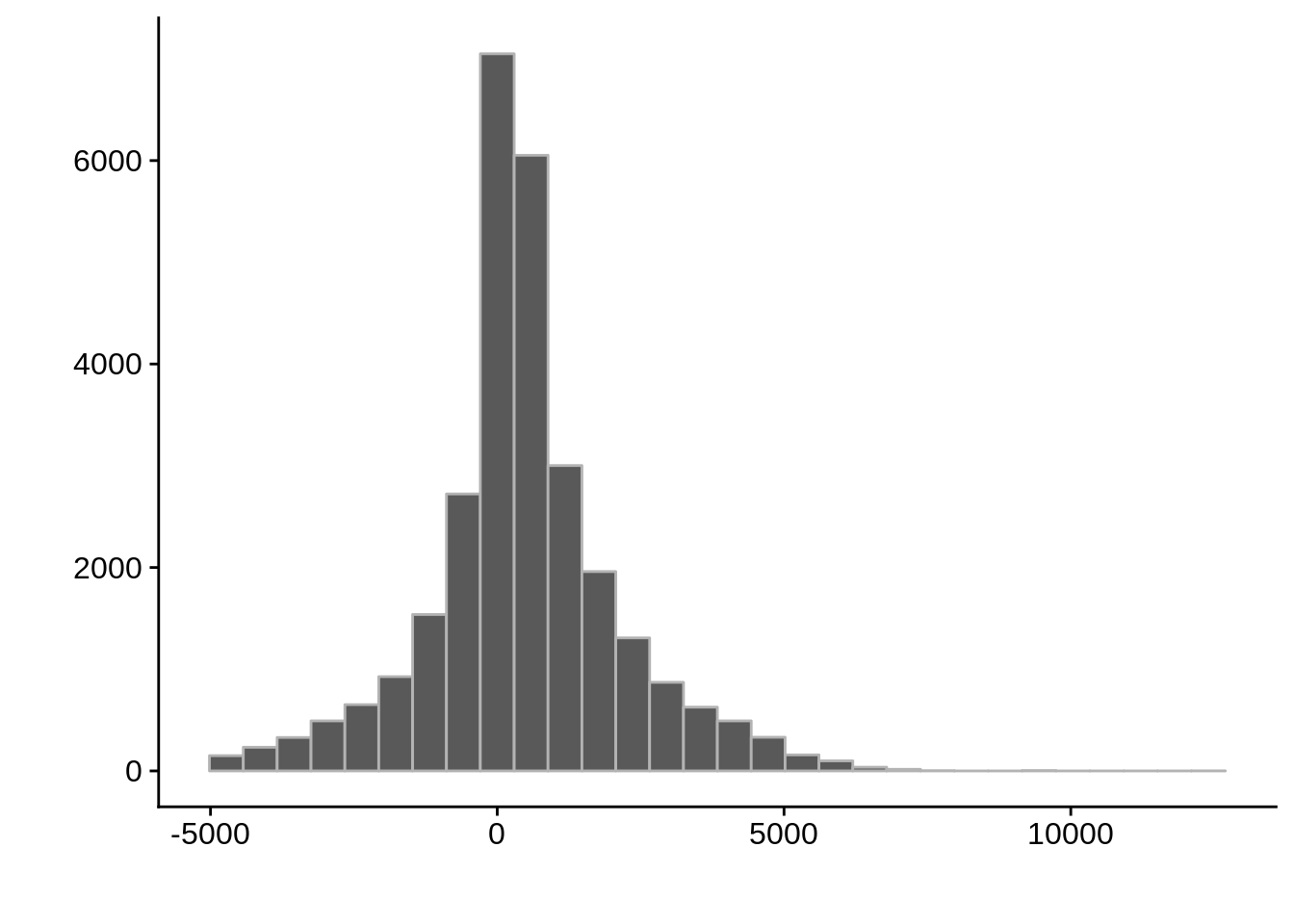
Expand here to see past versions of unnamed-chunk-9-2.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
AssignedFeat position start diff
Length:18268 Min. : 41434 Min. : 43094 Min. :-4967.0
Class :character 1st Qu.: 417702 1st Qu.: 417063 1st Qu.: -288.1
Mode :character Median : 810819 Median : 811370 Median : 295.0
Mean : 948485 Mean : 948096 Mean : 388.8
3rd Qu.:1286032 3rd Qu.:1286107 3rd Qu.: 1087.0
Max. :3207218 Max. :3207055 Max. :12155.0 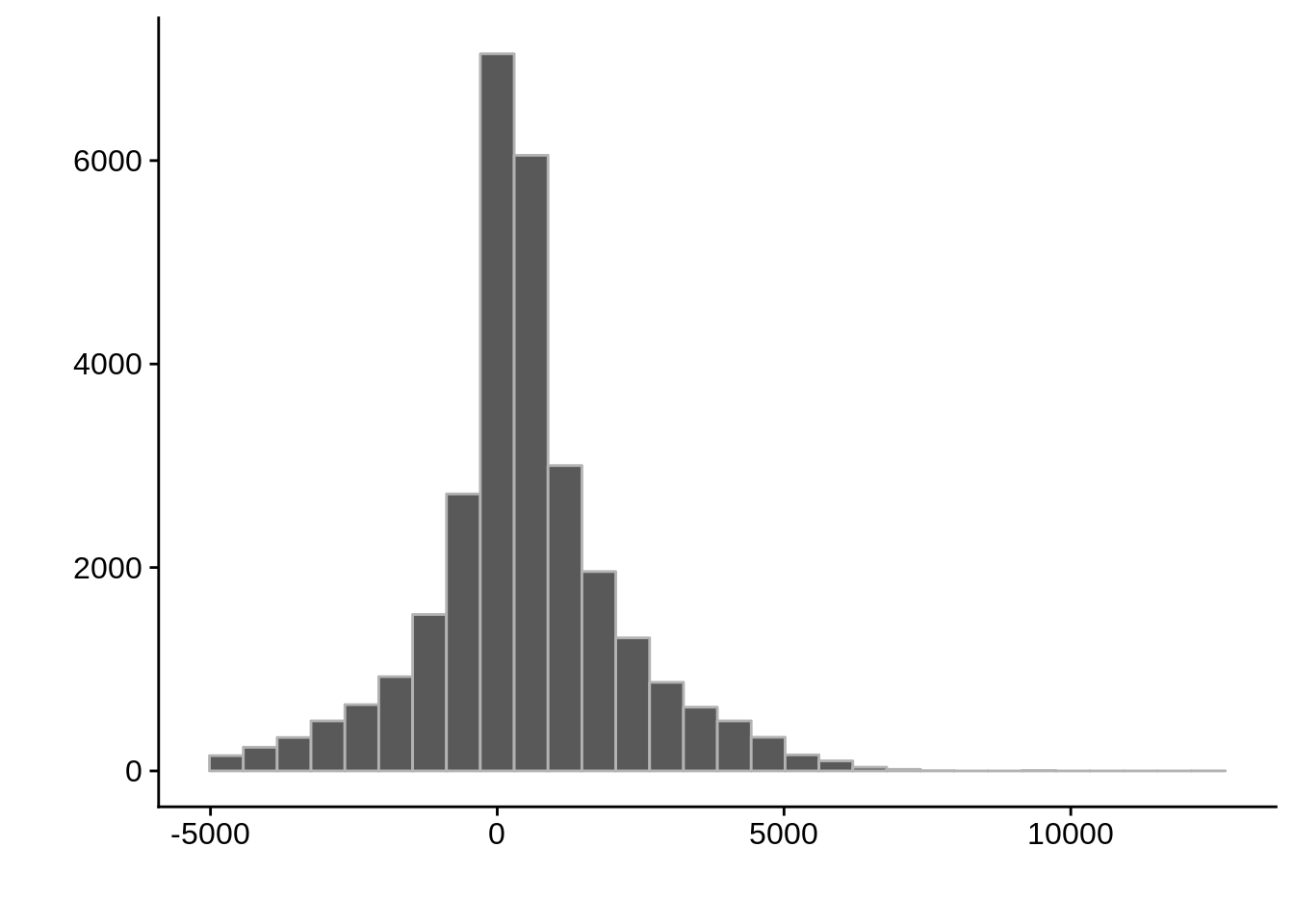
Expand here to see past versions of unnamed-chunk-9-3.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
AssignedFeat position start diff
Length:10086 Min. : 41434 Min. : 43094 Min. :-4957.5
Class :character 1st Qu.: 417660 1st Qu.: 417063 1st Qu.: -247.5
Mode :character Median : 811348 Median : 811370 Median : 291.0
Mean : 940319 Mean : 939934 Mean : 385.0
3rd Qu.:1284996 3rd Qu.:1284735 3rd Qu.: 1034.4
Max. :3207218 Max. :3207055 Max. : 6628.0 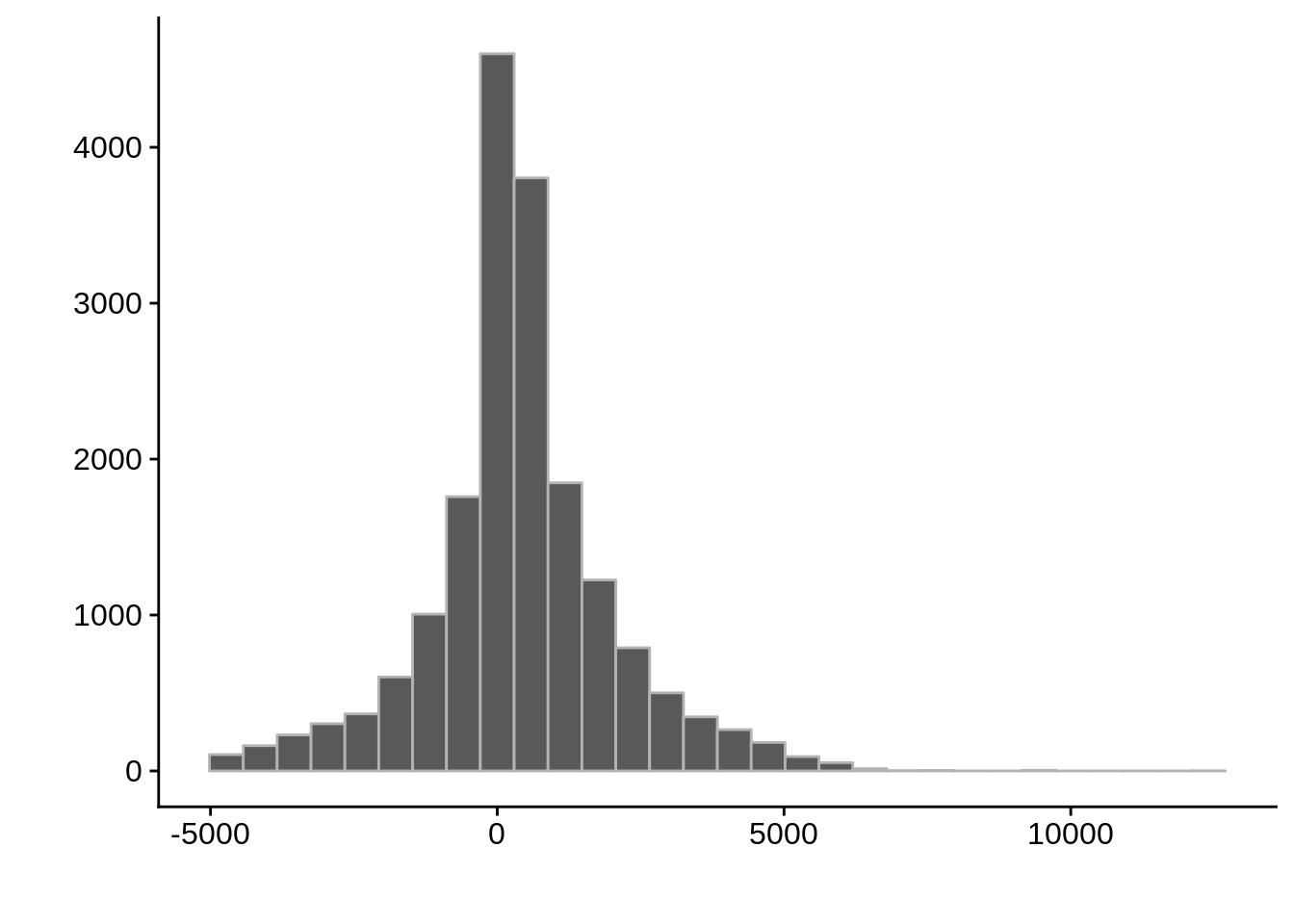
Expand here to see past versions of unnamed-chunk-9-4.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
AssignedFeat position start diff
Length:4644 Min. : 41434 Min. : 43094 Min. :-4948.5
Class :character 1st Qu.: 397119 1st Qu.: 397397 1st Qu.: -259.5
Mode :character Median : 766111 Median : 765911 Median : 244.8
Mean : 934627 Mean : 934337 Mean : 289.9
3rd Qu.:1295386 3rd Qu.:1293965 3rd Qu.: 878.0
Max. :3207218 Max. :3207055 Max. : 6100.0 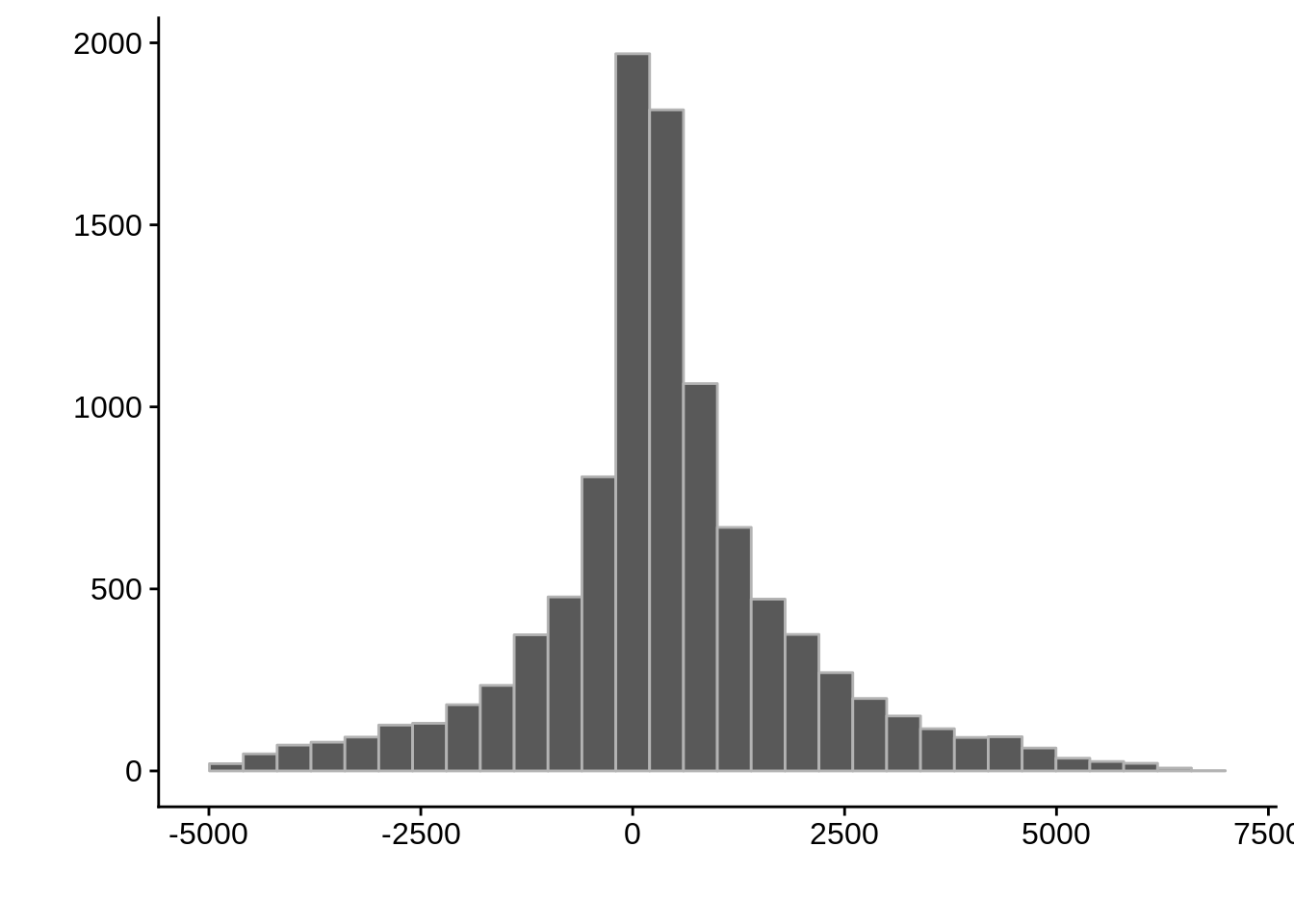
Expand here to see past versions of unnamed-chunk-9-5.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
AssignedFeat position start diff
Length:1665 Min. : 58482 Min. : 59727 Min. :-4948.5
Class :character 1st Qu.: 369837 1st Qu.: 368586 1st Qu.: -258.0
Mode :character Median : 792528 Median : 792937 Median : 227.5
Mean : 968848 Mean : 968578 Mean : 270.5
3rd Qu.:1357263 3rd Qu.:1357159 3rd Qu.: 845.0
Max. :2889297 Max. :2888660 Max. : 6100.0 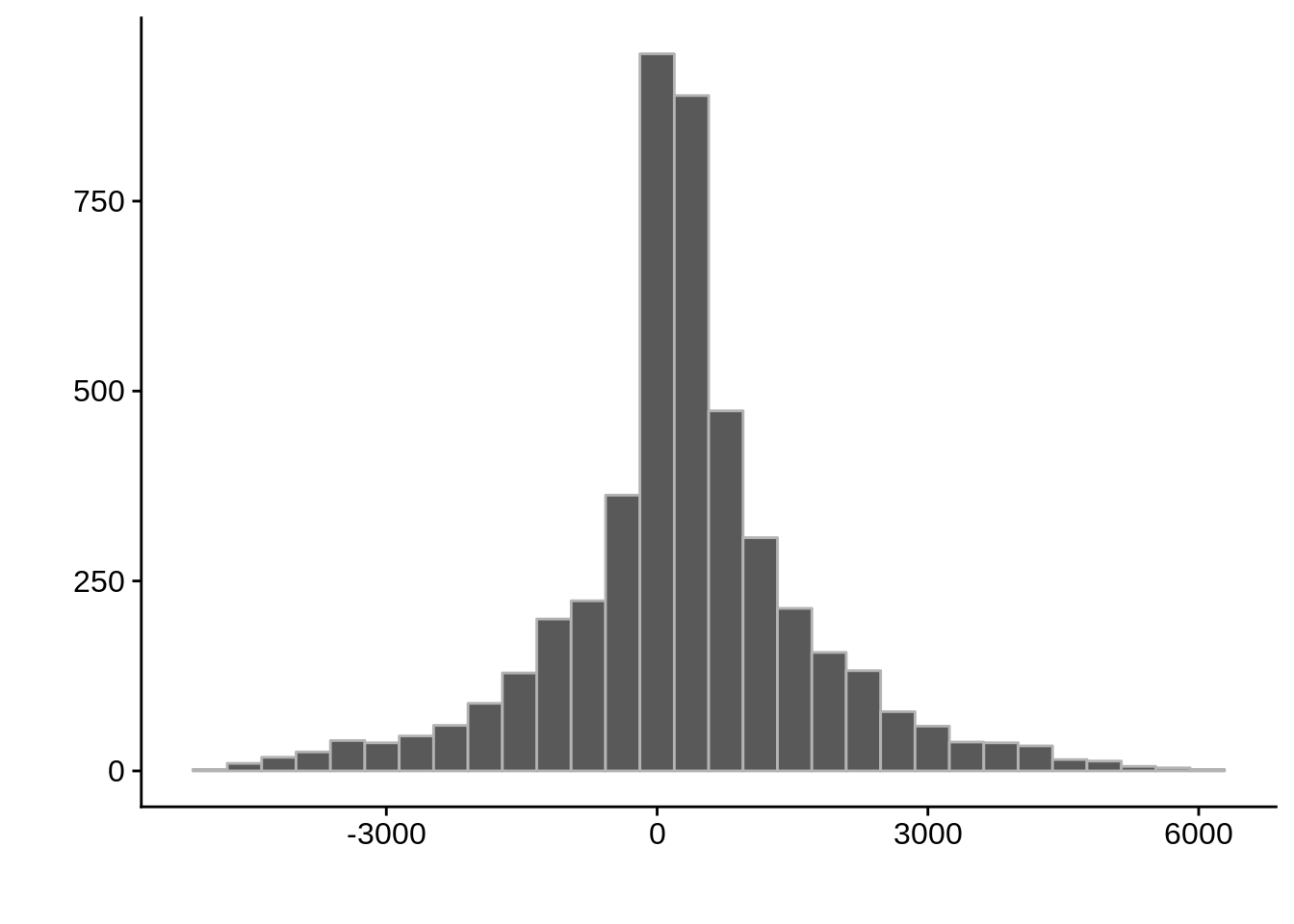
Expand here to see past versions of unnamed-chunk-9-6.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
AssignedFeat position start diff
Length:511 Min. : 83962 Min. : 83928 Min. :-4615.5
Class :character 1st Qu.: 319338 1st Qu.: 318727 1st Qu.: -286.2
Mode :character Median : 758262 Median : 758556 Median : 181.0
Mean : 923067 Mean : 922854 Mean : 212.9
3rd Qu.:1302403 3rd Qu.:1301308 3rd Qu.: 740.0
Max. :2889297 Max. :2888660 Max. : 4699.5 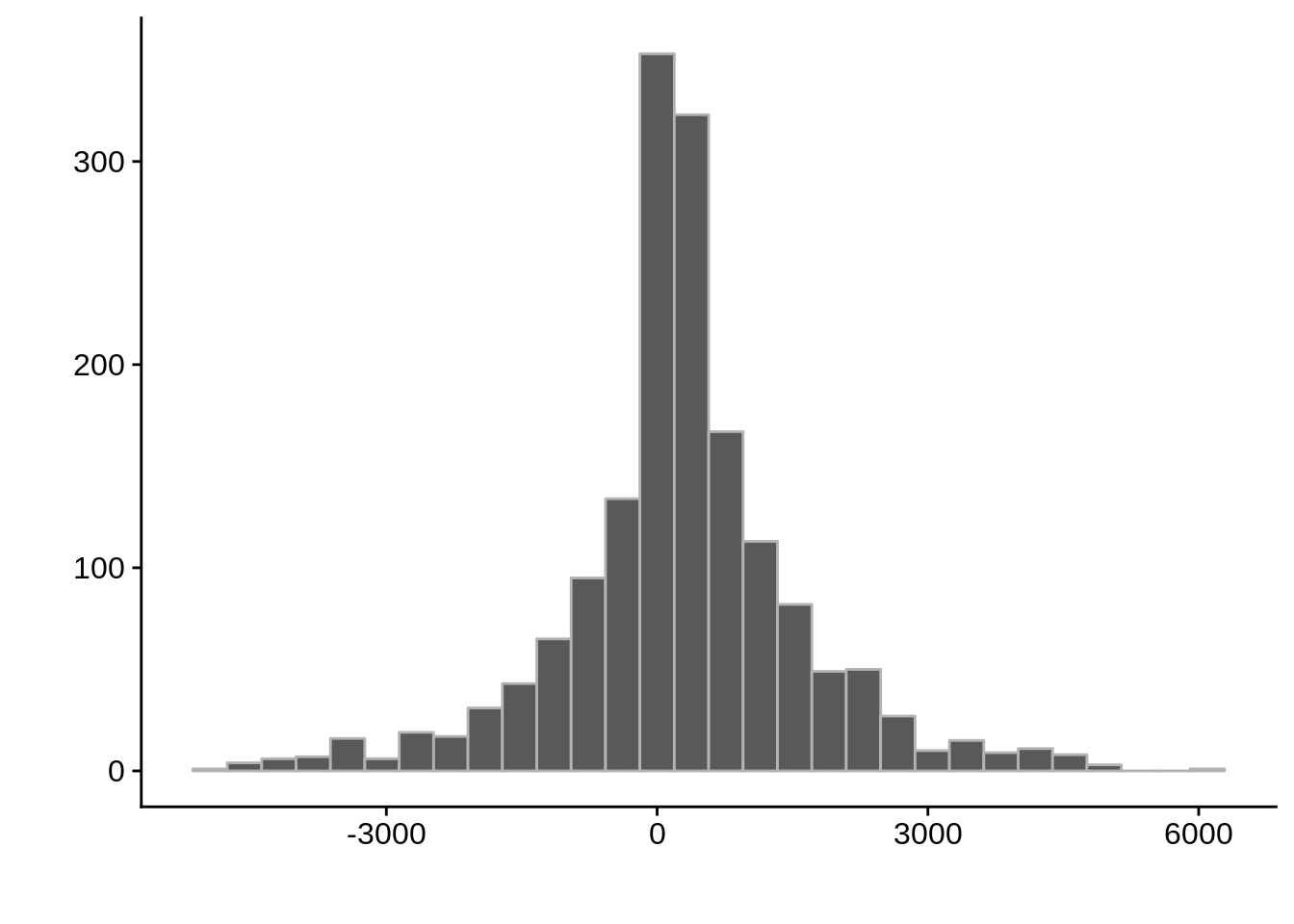
Expand here to see past versions of unnamed-chunk-9-7.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
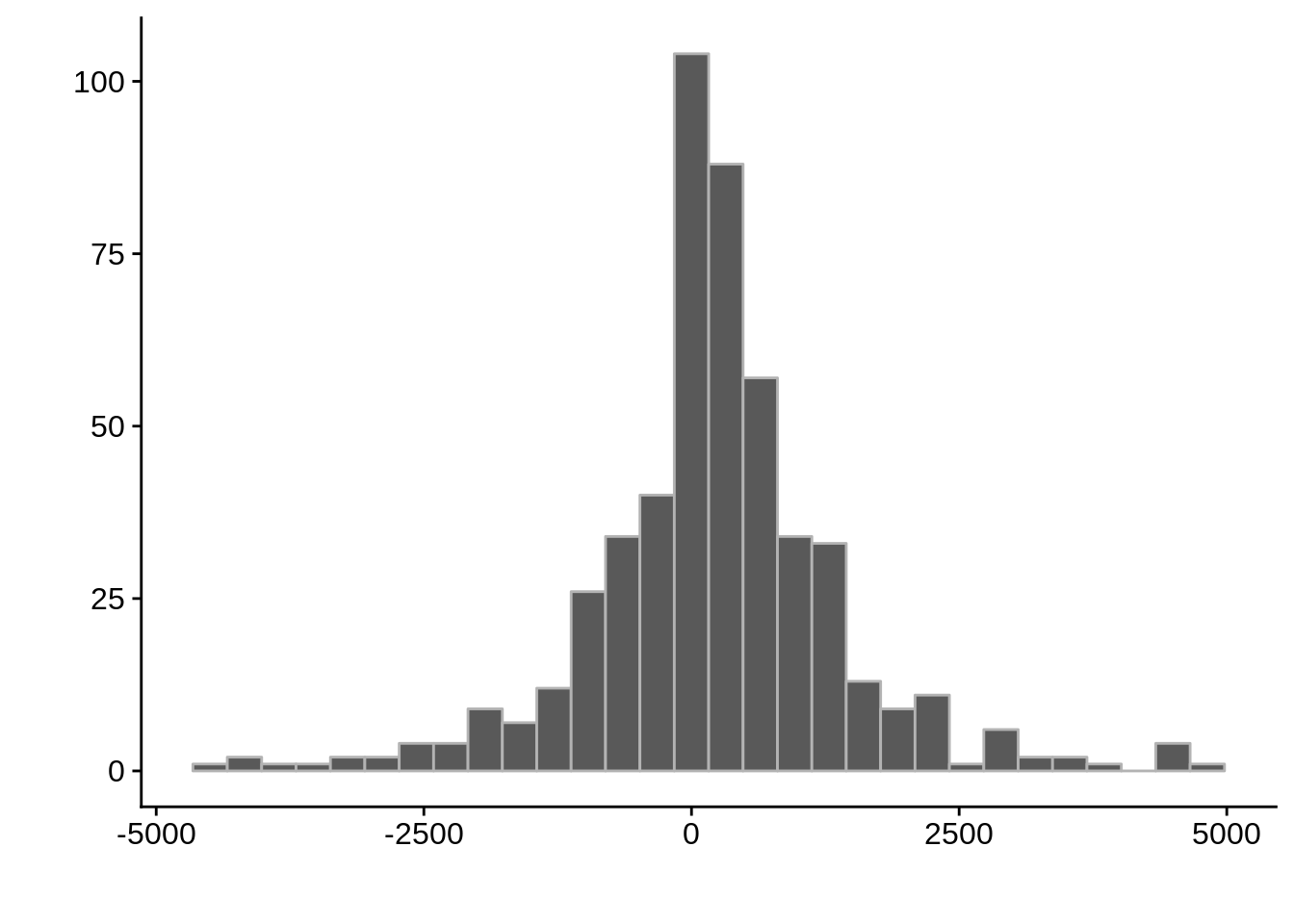
Expand here to see past versions of unnamed-chunk-9-8.png:
| Version | Author | Date |
|---|---|---|
| 1e6d9bb | Philipp Ross | 2018-09-24 |
Session information
R version 3.5.0 (2018-04-23)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Gentoo/Linux
Matrix products: default
BLAS: /usr/local/lib64/R/lib/libRblas.so
LAPACK: /usr/local/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] bindrcpp_0.2.2
[2] gdtools_0.1.7
[3] BSgenome.Pfalciparum.PlasmoDB.v24_1.0
[4] BSgenome_1.48.0
[5] rtracklayer_1.40.6
[6] Biostrings_2.48.0
[7] XVector_0.20.0
[8] GenomicRanges_1.32.7
[9] GenomeInfoDb_1.16.0
[10] org.Pf.plasmo.db_3.6.0
[11] AnnotationDbi_1.42.1
[12] IRanges_2.14.12
[13] S4Vectors_0.18.3
[14] Biobase_2.40.0
[15] BiocGenerics_0.26.0
[16] scales_1.0.0
[17] cowplot_0.9.3
[18] magrittr_1.5
[19] forcats_0.3.0
[20] stringr_1.3.1
[21] dplyr_0.7.6
[22] purrr_0.2.5
[23] readr_1.1.1
[24] tidyr_0.8.1
[25] tibble_1.4.2
[26] ggplot2_3.0.0
[27] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] nlme_3.1-137 bitops_1.0-6
[3] matrixStats_0.54.0 lubridate_1.7.4
[5] bit64_0.9-7 httr_1.3.1
[7] rprojroot_1.3-2 tools_3.5.0
[9] backports_1.1.2 R6_2.3.0
[11] DBI_1.0.0 lazyeval_0.2.1
[13] colorspace_1.3-2 withr_2.1.2
[15] tidyselect_0.2.4 bit_1.1-14
[17] compiler_3.5.0 git2r_0.23.0
[19] cli_1.0.1 rvest_0.3.2
[21] xml2_1.2.0 DelayedArray_0.6.6
[23] labeling_0.3 digest_0.6.17
[25] Rsamtools_1.32.3 svglite_1.2.1
[27] rmarkdown_1.10 R.utils_2.7.0
[29] pkgconfig_2.0.2 htmltools_0.3.6
[31] rlang_0.2.2 readxl_1.1.0
[33] rstudioapi_0.8 RSQLite_2.1.1
[35] bindr_0.1.1 jsonlite_1.5
[37] BiocParallel_1.14.2 R.oo_1.22.0
[39] RCurl_1.95-4.11 GenomeInfoDbData_1.1.0
[41] Matrix_1.2-14 Rcpp_0.12.19
[43] munsell_0.5.0 R.methodsS3_1.7.1
[45] stringi_1.2.4 whisker_0.3-2
[47] yaml_2.2.0 SummarizedExperiment_1.10.1
[49] zlibbioc_1.26.0 plyr_1.8.4
[51] grid_3.5.0 blob_1.1.1
[53] crayon_1.3.4 lattice_0.20-35
[55] haven_1.1.2 hms_0.4.2
[57] knitr_1.20 pillar_1.3.0
[59] XML_3.98-1.16 glue_1.3.0
[61] evaluate_0.11 modelr_0.1.2
[63] cellranger_1.1.0 gtable_0.2.0
[65] assertthat_0.2.0 broom_0.5.0
[67] GenomicAlignments_1.16.0 memoise_1.1.0
[69] workflowr_1.1.1
This reproducible R Markdown analysis was created with workflowr 1.1.1